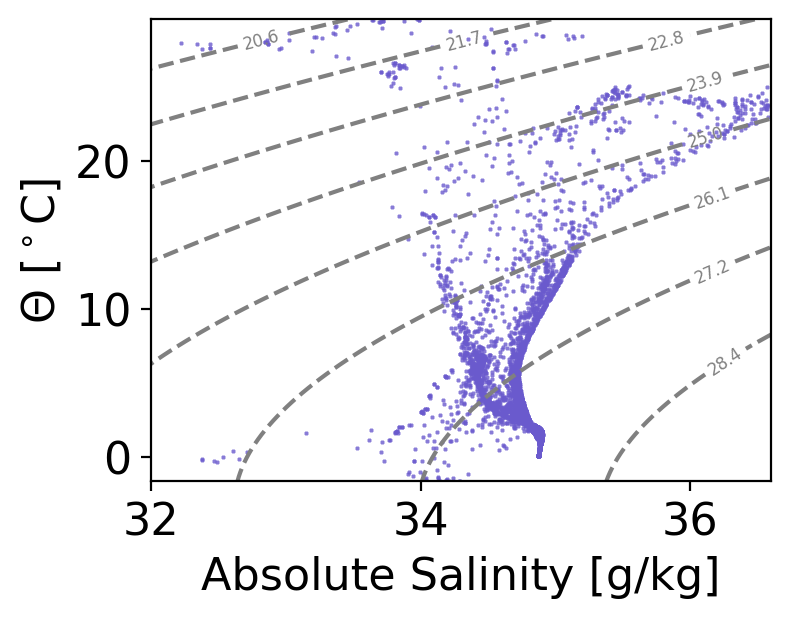
Explore Bottle File Data from CCHDO#
In this notebook we will plot data from a bottle file, collected on a repeat hydrographic section part of the GO-SHIP repeat hydrographic program.
All hydrographic data part of this progam are publicly avaiable and are archived at CCHDO. The section analyzed here (P18) is a meridonal transect in the eastern Pacific roughly along the 103\(^o\)W meridian. Section data is available at https://cchdo.ucsd.edu/cruise/33RO20161119. The netCDF file for the bottle data can be downloaded here.
import xarray as xr
import pandas as pd
import numpy as np
import datetime
import matplotlib.pyplot as plt
import matplotlib as mpl
import cartopy.crs as ccrs
import gsw
%matplotlib inline
plt.rcParams["font.size"] = 16
plt.rcParams["figure.facecolor"] = 'white'
import warnings
warnings.filterwarnings('ignore')
datapath = './data/'
# Load netCDF file locally as xarray Dataset
dd = xr.load_dataset(datapath+'p18_btl.nc')
dd
<xarray.Dataset> Size: 3MB
Dimensions: (N_PROF: 213, N_LEVELS: 24)
Coordinates:
expocode (N_PROF) object 2kB '33RO20161119' ......
station (N_PROF) object 2kB '1' '2' ... '212'
cast (N_PROF) int32 852B 3 2 1 1 1 ... 1 1 1 1
sample (N_PROF, N_LEVELS) object 41kB '24' .....
time (N_PROF) datetime64[ns] 2kB 2016-11-24...
latitude (N_PROF) float64 2kB 22.69 ... -68.07
longitude (N_PROF) float64 2kB -110.0 ... -95.0
pressure (N_PROF, N_LEVELS) float64 41kB 3.1 .....
Dimensions without coordinates: N_PROF, N_LEVELS
Data variables: (12/86)
section_id (N_PROF) object 2kB 'P18' 'P18' ... 'P18'
bottle_number (N_PROF, N_LEVELS) object 41kB '11122'...
bottle_number_qc (N_PROF, N_LEVELS) float32 20kB 2.0 .....
btm_depth (N_PROF) float64 2kB 2.619e+03 ... 4.4...
ctd_temperature (N_PROF, N_LEVELS) float64 41kB 27.88 ...
ctd_salinity (N_PROF, N_LEVELS) float64 41kB 34.53 ...
... ...
n2_argon_ratio_unstripped_error (N_PROF, N_LEVELS) float64 41kB nan .....
d15n_n2 (N_PROF, N_LEVELS) float64 41kB nan .....
d15n_n2_qc (N_PROF, N_LEVELS) float32 20kB nan .....
d15n_n2_error (N_PROF, N_LEVELS) float64 41kB nan .....
profile_type (N_PROF) object 2kB 'B' 'B' ... 'B' 'B'
geometry_container float64 8B nan
Attributes:
Conventions: CF-1.8 CCHDO-1.0
cchdo_software_version: hydro 1.0.2.8
cchdo_parameters_version: params 2024.4.0
comments: BOTTLE,20230606CCHSIOCBG\n Merged parameters: ...
featureType: profile- N_PROF: 213
- N_LEVELS: 24
- expocode(N_PROF)object'33RO20161119' ... '33RO20161119'
- whp_name :
- EXPOCODE
- geometry :
- geometry_container
array(['33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', ... '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119', '33RO20161119'], dtype=object) - station(N_PROF)object'1' '2' '3' ... '210' '211' '212'
- whp_name :
- STNNBR
- geometry :
- geometry_container
array(['1', '2', '3', '5', '6', '4', '7', '8', '9', '10', '11', '12', '13', '14', '15', '16', '17', '18', '19', '20', '21', '22', '23', '24', '25', '26', '27', '28', '29', '30', '31', '32', '33', '34', '35', '36', '37', '38', '39', '40', '41', '42', '43', '44', '45', '46', '47', '48', '49', '50', '51', '52', '53', '54', '55', '56', '57', '58', '59', '60', '61', '62', '63', '64', '65', '66', '67', '68', '69', '70', '71', '72', '73', '74', '75', '76', '77', '78', '79', '80', '81', '82', '83', '84', '85', '86', '87', '88', '89', '90', '91', '92', '93', '94', '95', '96', '97', '98', '99', '100', '101', '102', '103', '104', '105', '106', '107', '108', '109', '110', '111', '112', '113', '114', '115', '116', '120', '117', '118', '119', '121', '122', '123', '124', '125', '126', '127', '128', '129', '130', '131', '132', '133', '134', '135', '136', '137', '138', '139', '140', '141', '142', '143', '144', '145', '146', '147', '148', '149', '150', '151', '152', '153', '154', '155', '156', '157', '158', '159', '160', '161', '162', '163', '164', '165', '166', '167', '168', '169', '170', '171', '172', '173', '174', '175', '176', '177', '178', '179', '180', '181', '182', '183', '184', '185', '186', '187', '188', '189', '190', '191', '192', '193', '194', '195', '196', '197', '198', '199', '200', '201', '202', '203', '204', '205', '206', '206', '207', '208', '209', '210', '211', '212'], dtype=object) - cast(N_PROF)int323 2 1 1 1 1 1 1 ... 1 2 1 1 1 1 1 1
- whp_name :
- CASTNO
- geometry :
- geometry_container
array([3, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1], dtype=int32) - sample(N_PROF, N_LEVELS)object'24' '23' '22' '21' ... '3' '2' '1'
- whp_name :
- SAMPNO
array([['24', '23', '22', ..., '3', '2', '1'], ['10', '11', '9', ..., '', '', ''], ['23', '24', '22', ..., '3', '2', '1'], ..., ['24', '23', '22', ..., '3', '2', '1'], ['24', '23', '22', ..., '3', '2', '1'], ['24', '23', '22', ..., '3', '2', '1']], dtype=object) - time(N_PROF)datetime64[ns]2016-11-24T14:17:00 ... 2017-01-...
- standard_name :
- time
- whp_name :
- ['DATE', 'TIME']
- resolution :
- 0.0006944444444444444
- axis :
- T
- geometry :
- geometry_container
array(['2016-11-24T14:17:00.000000000', '2016-11-24T18:54:00.000000000', '2016-11-24T21:37:00.000000000', '2016-11-25T07:46:00.000000000', '2016-11-25T13:09:00.000000000', '2016-11-25T15:50:00.000000000', '2016-11-25T18:44:00.000000000', '2016-11-26T00:13:00.000000000', '2016-11-26T06:02:00.000000000', '2016-11-26T11:27:00.000000256', '2016-11-26T17:08:00.000000000', '2016-11-26T22:44:00.000000000', '2016-11-27T04:32:00.000000000', '2016-11-27T10:34:00.000000256', '2016-11-27T16:25:00.000000000', '2016-11-27T22:19:00.000000000', '2016-11-28T04:09:59.999999744', '2016-11-28T10:27:00.000000000', '2016-11-28T17:02:00.000000000', '2016-11-29T03:49:00.000000256', '2016-11-29T09:48:00.000000000', '2016-11-29T15:34:00.000000000', '2016-11-29T21:47:00.000000000', '2016-11-30T03:52:00.000000000', '2016-11-30T10:03:00.000000000', '2016-11-30T16:03:00.000000000', '2016-11-30T23:12:00.000000000', '2016-12-01T04:53:00.000000000', '2016-12-01T10:28:00.000000000', '2016-12-01T16:08:00.000000000', '2016-12-01T22:11:00.000000000', '2016-12-02T04:13:00.000000256', '2016-12-02T10:20:00.000000000', '2016-12-02T16:34:00.000000256', '2016-12-02T22:17:59.999999744', '2016-12-03T03:52:00.000000000', '2016-12-03T09:24:59.999999744', '2016-12-03T15:07:00.000000000', '2016-12-03T21:12:00.000000256', '2016-12-04T03:08:00.000000000', ... '2017-01-16T19:17:59.999999744', '2017-01-17T01:56:00.000000000', '2017-01-17T08:31:00.000000000', '2017-01-17T15:29:00.000000000', '2017-01-17T22:09:00.000000000', '2017-01-18T04:46:59.999999744', '2017-01-18T11:21:00.000000000', '2017-01-18T17:46:00.000000000', '2017-01-19T00:43:00.000000000', '2017-01-19T08:49:00.000000000', '2017-01-19T16:11:00.000000000', '2017-01-19T23:06:00.000000000', '2017-01-20T06:03:00.000000000', '2017-01-20T13:07:00.000000000', '2017-01-20T19:57:00.000000000', '2017-01-21T03:04:00.000000256', '2017-01-21T10:06:00.000000000', '2017-01-21T17:31:00.000000000', '2017-01-22T00:31:00.000000000', '2017-01-22T07:25:59.999999744', '2017-01-22T14:23:00.000000000', '2017-01-22T21:08:00.000000000', '2017-01-23T04:09:59.999999744', '2017-01-23T11:24:00.000000000', '2017-01-23T18:19:00.000000000', '2017-01-24T01:08:00.000000000', '2017-01-24T08:08:00.000000000', '2017-01-24T15:00:00.000000000', '2017-01-25T00:57:00.000000256', '2017-01-25T09:38:00.000000000', '2017-01-25T18:21:00.000000000', '2017-01-26T01:57:00.000000000', '2017-01-26T01:57:00.000000000', '2017-01-26T16:00:00.000000000', '2017-01-26T21:32:00.000000000', '2017-01-27T07:31:00.000000000', '2017-01-27T18:04:00.000000256', '2017-01-27T23:43:00.000000256', '2017-01-29T02:45:00.000000000'], dtype='datetime64[ns]') - latitude(N_PROF)float6422.69 22.87 22.77 ... -70.0 -68.07
- whp_name :
- LATITUDE
- standard_name :
- latitude
- units :
- degree_north
- C_format :
- %.4f
- C_format_source :
- input_file
- axis :
- Y
array([ 2.26884e+01, 2.28696e+01, 2.27705e+01, 2.20021e+01, 2.14988e+01, 2.25017e+01, 2.10000e+01, 2.05008e+01, 2.00002e+01, 1.94999e+01, 1.89999e+01, 1.85007e+01, 1.80002e+01, 1.74997e+01, 1.69999e+01, 1.65005e+01, 1.60014e+01, 1.55007e+01, 1.49998e+01, 1.44988e+01, 1.40000e+01, 1.34997e+01, 1.30001e+01, 1.25001e+01, 1.19998e+01, 1.14997e+01, 1.10009e+01, 1.05006e+01, 1.00001e+01, 9.49960e+00, 8.99980e+00, 8.49990e+00, 7.99980e+00, 7.50000e+00, 7.00010e+00, 6.49910e+00, 5.99990e+00, 5.50000e+00, 4.99980e+00, 4.50010e+00, 4.00000e+00, 3.50020e+00, 3.00000e+00, 2.75020e+00, 2.50170e+00, 2.25010e+00, 2.00030e+00, 1.75000e+00, 1.50020e+00, 1.25070e+00, 1.00000e+00, 7.50000e-01, 5.01200e-01, 2.50300e-01, 3.80000e-03, -2.45500e-01, -4.95700e-01, -7.42800e-01, -9.99700e-01, -1.24880e+00, -1.49990e+00, -1.74940e+00, -1.99970e+00, -2.25010e+00, -2.50000e+00, -2.74970e+00, -2.99960e+00, -3.50020e+00, -3.99980e+00, -4.49980e+00, -4.99980e+00, -5.29020e+00, -5.59010e+00, -5.88020e+00, -6.17990e+00, -6.46980e+00, -6.76010e+00, -7.06000e+00, -7.35000e+00, -7.64980e+00, ... -3.44999e+01, -3.50000e+01, -3.55010e+01, -3.60003e+01, -3.64996e+01, -3.70006e+01, -3.75003e+01, -3.79998e+01, -3.84995e+01, -3.89999e+01, -3.95005e+01, -4.00000e+01, -4.05001e+01, -4.09985e+01, -4.15007e+01, -4.20002e+01, -4.25009e+01, -4.29996e+01, -4.35005e+01, -4.40000e+01, -4.45003e+01, -4.50017e+01, -4.55000e+01, -4.60001e+01, -4.65001e+01, -4.70003e+01, -4.75000e+01, -4.80000e+01, -4.85003e+01, -4.90001e+01, -4.94998e+01, -5.00000e+01, -5.04991e+01, -5.10001e+01, -5.15000e+01, -5.20004e+01, -5.24998e+01, -5.30001e+01, -5.34998e+01, -5.40001e+01, -5.44999e+01, -5.50001e+01, -5.55005e+01, -5.60002e+01, -5.65000e+01, -5.70000e+01, -5.75005e+01, -5.80003e+01, -5.85017e+01, -5.89999e+01, -5.95003e+01, -6.00006e+01, -6.05002e+01, -6.09995e+01, -6.14999e+01, -6.20001e+01, -6.25025e+01, -6.29999e+01, -6.35401e+01, -6.39997e+01, -6.45006e+01, -6.49999e+01, -6.55001e+01, -6.59887e+01, -6.65003e+01, -6.69973e+01, -6.75004e+01, -6.80000e+01, -6.85001e+01, -6.90009e+01, -6.90009e+01, -6.95047e+01, -6.96067e+01, -6.96910e+01, -6.99009e+01, -7.00012e+01, -6.80668e+01]) - longitude(N_PROF)float64-110.0 -110.0 ... -100.2 -95.0
- whp_name :
- LONGITUDE
- standard_name :
- longitude
- units :
- degree_east
- C_format :
- %.4f
- C_format_source :
- input_file
- axis :
- X
array([-110.0001, -110.0003, -110. , -109.9999, -109.9999, -109.9992, -110.0003, -109.999 , -110. , -109.9986, -110. , -110.0006, -110.0004, -110.0006, -110.0003, -110.0006, -109.9998, -110.0002, -109.9998, -109.9998, -110.0001, -110.0001, -110.0007, -109.9996, -110.0006, -110.0004, -109.9986, -110.0008, -109.9998, -110.0001, -109.9999, -109.9994, -110. , -109.9996, -110. , -109.9999, -109.9999, -109.9998, -110.0005, -109.9994, -110. , -110. , -110.0001, -110.001 , -109.9998, -110.0001, -110.0001, -110. , -110.0001, -109.9999, -110. , -110.0001, -110.0003, -109.9979, -110.0004, -109.9969, -110.0015, -109.9994, -110.0002, -110.0009, -110.0001, -110.001 , -109.9994, -109.9999, -109.9999, -110. , -109.9998, -110. , -110.0002, -110. , -109.9999, -109.59 , -109.1799, -108.7601, -108.3499, -107.94 , -107.5294, -107.1202, -106.71 , -106.2905, -105.8801, -105.4701, -105.06 , -104.6502, -104.24 , -103.8201, -103.41 , -102.9999, -103. , -103.0002, -103.0001, -103.0004, -103. , -103.0001, -103. , -103.0002, -103.0001, -102.9999, -103. , -102.9995, -103. , -102.9995, -103. , -102.9997, -103.0012, -102.9995, -103. , -103. , -102.9974, -103.0001, -103. , -103.0003, -102.9973, -103.0001, -103.0006, -103.0002, -101.4998, -102.3335, -101.5005, -101.4999, -101.4997, -101.4992, -101.5009, -101.5001, -102.3333, -103.0075, -102.9991, -102.9998, -103.0003, -102.9993, -103. , -102.9993, -103. , -102.9989, -102.9987, -103.0003, -103. , -103.0002, -103.0027, -103. , -103.0013, -102.9984, -102.9994, -102.9999, -103.0006, -103. , -103.0044, -102.9992, -103.0003, -103.0023, -103.0005, -103.0002, -102.9977, -102.9996, -102.9995, -102.9999, -103.0004, -103.0006, -102.9993, -103.0001, -103. , -102.9995, -103.0001, -102.9997, -102.999 , -102.9998, -103.0004, -102.9992, -103.0001, -102.9988, -103.0002, -102.9999, -102.9998, -103.0001, -103.0004, -103.0006, -102.9999, -102.9995, -103.0007, -103. , -102.9997, -102.9996, -102.9978, -103. , -102.9976, -103. , -102.9989, -103.0002, -102.9989, -103.0008, -102.9996, -102.9965, -102.9984, -102.9995, -102.9811, -102.9998, -103.0002, -103.0003, -103. , -102.9979, -103.0006, -103.0002, -103.023 , -102.9991, -103.0001, -102.9937, -102.9937, -102.9941, -103.0221, -102.0269, -100.6698, -100.2423, -94.9996]) - pressure(N_PROF, N_LEVELS)float643.1 25.0 ... 4.163e+03 4.52e+03
- whp_name :
- CTDPRS
- whp_unit :
- DBAR
- standard_name :
- sea_water_pressure
- units :
- dbar
- C_format :
- %.1f
- C_format_source :
- input_file
- axis :
- Z
- positive :
- down
array([[3.1000e+00, 2.5000e+01, 6.0100e+01, ..., 2.1001e+03, 2.3506e+03, 2.6416e+03], [3.3000e+00, 3.4000e+00, 3.2700e+01, ..., nan, nan, nan], [3.0000e+00, 3.1000e+00, 2.0300e+01, ..., 9.6500e+02, 1.0955e+03, 1.2560e+03], ..., [2.6000e+00, 2.5400e+01, 6.0500e+01, ..., 3.5510e+03, 3.8565e+03, 4.1616e+03], [3.1000e+00, 3.0300e+01, 7.0100e+01, ..., 3.6552e+03, 3.9116e+03, 4.1667e+03], [3.3000e+00, 1.7900e+01, 3.4100e+01, ..., 3.8052e+03, 4.1630e+03, 4.5196e+03]])
- section_id(N_PROF)object'P18' 'P18' 'P18' ... 'P18' 'P18'
- whp_name :
- SECT_ID
- geometry :
- geometry_container
array(['P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18', 'P18'], dtype=object) - bottle_number(N_PROF, N_LEVELS)object'11122' '11121' ... '11101' '11010'
- whp_name :
- BTLNBR
- ancillary_variables :
- bottle_number_qc
array([['11122', '11121', '11120', ..., '11102', '11101', '11010'], ['11109', '11110', '11108', ..., '', '', ''], ['11121', '11122', '11120', ..., '11102', '11101', '11010'], ..., ['11122', '11121', '11120', ..., '11102', '11101', '11010'], ['11122', '11121', '11120', ..., '11102', '11101', '11010'], ['11122', '11121', '11120', ..., '11102', '11101', '11010']], dtype=object) - bottle_number_qc(N_PROF, N_LEVELS)float322.0 2.0 2.0 2.0 ... 2.0 2.0 2.0 2.0
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned bottle_information_unavailable no_problems_noted leaking did_not_trip_correctly not_reported significant_discrepancy_in_measured_values_between_gerard_and_niskin_bottles unknown_problem pair_did_not_trip_correctly_note_that_the_niskin_bottle_can_trip_at_an_unplanned_depth_while_the_gerard_trips_correctly_and_vice_versa samples_not_drawn_from_this_bottle
- conventions :
- WOCESAMPLE - WOCE Quality Codes for the sampling device itself
array([[ 2., 2., 2., ..., 2., 2., 2.], [ 2., 2., 2., ..., nan, nan, nan], [ 2., 2., 2., ..., 2., 2., 2.], ..., [ 2., 2., 2., ..., 2., 2., 2.], [ 2., 2., 2., ..., 2., 2., 2.], [ 2., 2., 2., ..., 2., 2., 2.]], dtype=float32) - btm_depth(N_PROF)float642.619e+03 2.619e+03 ... 4.429e+03
- whp_name :
- DEPTH
- whp_unit :
- METERS
- standard_name :
- sea_floor_depth_below_sea_surface
- units :
- meters
- C_format :
- %.0f
- C_format_source :
- input_file
array([2619., 2619., 1250., 3157., 3196., 3047., 3250., 3110., 2564., 3259., 3369., 3441., 3281., 3420., 3511., 3410., 3295., 3770., 3785., 3543., 3184., 3792., 3267., 3751., 3096., 3755., 3724., 3353., 3310., 3595., 4117., 3888., 3623., 3814., 3755., 3591., 3693., 4003., 3915., 3918., 3877., 3830., 3892., 3837., 3741., 3792., 3749., 3727., 3772., 3721., 3810., 3713., 3800., 3812., 3818., 3737., 3908., 3925., 3983., 3921., 3850., 3904., 3912., 3903., 3931., 3809., 3765., 3946., 3722., 3605., 3617., 3510., 3444., 3395., 3208., 3095., 2919., 3144., 3483., 3555., 3494., 3562., 3743., 3635., 3924., 4127., 4014., 4564., 4697., 4279., 4093., 4359., 4186., 4356., 4120., 4223., 3966., 4160., 3749., 3745., 3087., 3925., 4031., 4173., 4061., 4111., 4126., 4215., 4054., 4070., 3966., 3915., 4023., 3986., 3927., 3959., 3331., 3642., 3558., 3540., 3467., 2400., 3184., 3402., 3587., 3366., 3470., 3616., 3498., 3461., 3531., 3658., 3585., 3665., 3592., 3626., 3424., 3519., 3001., 3806., 3515., 4012., 3477., 4138., 3942., 3736., 3936., 3964., 3896., 3976., 3727., 3906., 3792., 3816., 3759., 3942., 3808., 3776., 3693., 3915., 3857., 4059., 4239., 4148., 4162., 4240., 4109., 4127., 4220., 4386., 4061., 4289., 4433., 4136., 4270., 4303., 4101., 4605., 4659., 4455., 4327., 4183., 4421., 4542., 4708., 4648., 4520., 4961., 5275., 4977., 5148., 5078., 5057., 5026., 5017., 5001., 4964., 4942., 4882., 4840., 4785., 4723., 4633., 4487., 4306., 4063., 4063., 4088., 4146., 3936., 4093., 4099., 4429.]) - ctd_temperature(N_PROF, N_LEVELS)float6427.88 27.89 21.33 ... 0.3916 0.3832
- whp_name :
- CTDTMP
- whp_unit :
- ITS-90
- standard_name :
- sea_water_temperature
- units :
- degC
- reference_scale :
- ITS-90
- C_format :
- %.4f
- C_format_source :
- input_file
array([[27.8825, 27.893 , 21.327 , ..., 2.0198, 1.8914, 1.8216], [26.2906, 26.2932, 25.1281, ..., nan, nan, nan], [27.604 , 27.5713, 27.3301, ..., 4.5361, 4.1188, 3.5487], ..., [-0.1632, -1.038 , -1.6522, ..., 0.4531, 0.4338, 0.3607], [-0.3174, -1.5671, -1.6854, ..., 0.4106, 0.3789, 0.3511], [ 1.605 , 1.7837, -0.3206, ..., 0.4289, 0.3916, 0.3832]]) - ctd_salinity(N_PROF, N_LEVELS)float6434.53 34.52 34.18 ... 34.7 34.7
- whp_name :
- CTDSAL
- whp_unit :
- PSS-78
- standard_name :
- sea_water_practical_salinity
- units :
- 1
- reference_scale :
- PSS-78
- C_format :
- %.4f
- C_format_source :
- input_file
- ancillary_variables :
- ctd_salinity_qc
array([[34.5273, 34.5202, 34.1838, ..., 34.6435, 34.6537, 34.6618], [34.376 , 34.3765, 34.1194, ..., nan, nan, nan], [34.4895, 34.4933, 34.4815, ..., 34.5266, 34.5466, 34.5695], ..., [32.499 , 33.7559, 34.0486, ..., 34.7046, 34.7039, 34.7011], [32.3107, 33.976 , 34.1209, ..., 34.7036, 34.7026, 34.7008], [32.9924, 33.4734, 33.5649, ..., 34.7029, 34.7018, 34.7002]]) - ctd_salinity_qc(N_PROF, N_LEVELS)float322.0 2.0 2.0 2.0 ... 2.0 2.0 2.0 2.0
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 9]
- flag_meanings :
- no_flag_assigned not_calibrated acceptable_measurement questionable_measurement bad_measurement not_reported interpolated_over_a_pressure_interval_larger_than_2_dbar despiked not_sampled
- conventions :
- WOCECTD - WOCE Quality Codes for CTD instrument measurements
array([[ 2., 2., 2., ..., 2., 2., 2.], [ 2., 2., 2., ..., nan, nan, nan], [ 2., 2., 2., ..., 2., 2., 2.], ..., [ 2., 2., 2., ..., 2., 2., 2.], [ 2., 2., 2., ..., 2., 2., 2.], [ 2., 2., 2., ..., 2., 2., 2.]], dtype=float32) - bottle_salinity(N_PROF, N_LEVELS)float64nan nan 34.25 ... 34.7 34.71 34.7
- whp_name :
- SALNTY
- whp_unit :
- PSS-78
- standard_name :
- sea_water_practical_salinity
- units :
- 1
- reference_scale :
- PSS-78
- C_format :
- %.4f
- C_format_source :
- input_file
- ancillary_variables :
- bottle_salinity_qc
array([[ nan, nan, 34.2529, ..., 34.6469, 34.667 , 34.665 ], [34.3628, nan, 34.102 , ..., nan, nan, nan], [ nan, nan, 34.4862, ..., 34.5249, 34.5453, 34.5695], ..., [32.5343, 33.7679, 34.0479, ..., 34.703 , 34.7022, 34.7011], [32.3543, 33.9789, 34.1215, ..., 34.7022, 34.7012, 34.6994], [32.9924, 33.4775, 33.5661, ..., 34.7018, 34.7061, 34.7007]]) - bottle_salinity_qc(N_PROF, N_LEVELS)float32nan nan 2.0 2.0 ... 2.0 2.0 2.0 2.0
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, nan, 2., ..., 2., 4., 2.], [ 2., nan, 2., ..., nan, nan, nan], [nan, nan, 2., ..., 2., 2., 2.], ..., [ 2., 2., 2., ..., 2., 2., 2.], [ 2., 2., 2., ..., 2., 2., 2.], [ 2., 2., 2., ..., 2., 2., 2.]], dtype=float32) - ctd_oxygen(N_PROF, N_LEVELS)float64194.3 194.8 219.4 ... 212.3 213.5
- whp_name :
- CTDOXY
- whp_unit :
- UMOL/KG
- standard_name :
- moles_of_oxygen_per_unit_mass_in_sea_water
- units :
- umol/kg
- C_format :
- %.1f
- C_format_source :
- input_file
- ancillary_variables :
- ctd_oxygen_qc
array([[194.3, 194.8, 219.4, ..., 85.7, 93.9, 99.1], [196.3, 196.3, 207.2, ..., nan, nan, nan], [122.2, 139.4, 197.2, ..., 11.8, 17.5, 28.9], ..., [349.9, 347.5, 316.7, ..., 210. , 210.9, 213. ], [350. , 320.5, 297.7, ..., 211.3, 212.1, 213.1], [335.1, 338.9, 368.2, ..., 211.4, 212.3, 213.5]]) - ctd_oxygen_qc(N_PROF, N_LEVELS)float322.0 2.0 2.0 2.0 ... 2.0 2.0 2.0 2.0
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 9]
- flag_meanings :
- no_flag_assigned not_calibrated acceptable_measurement questionable_measurement bad_measurement not_reported interpolated_over_a_pressure_interval_larger_than_2_dbar despiked not_sampled
- conventions :
- WOCECTD - WOCE Quality Codes for CTD instrument measurements
array([[ 2., 2., 2., ..., 2., 2., 4.], [ 2., 2., 2., ..., nan, nan, nan], [ 2., 2., 2., ..., 2., 2., 2.], ..., [ 2., 2., 2., ..., 2., 2., 2.], [ 2., 2., 2., ..., 2., 2., 2.], [ 2., 2., 2., ..., 2., 2., 2.]], dtype=float32) - oxygen(N_PROF, N_LEVELS)float64nan 196.1 214.4 ... 212.0 212.8
- whp_name :
- OXYGEN
- whp_unit :
- UMOL/KG
- standard_name :
- moles_of_oxygen_per_unit_mass_in_sea_water
- units :
- umol/kg
- C_format :
- %.1f
- C_format_source :
- input_file
- ancillary_variables :
- oxygen_qc
array([[ nan, 196.1, 214.4, ..., 88.9, 96.5, 104.9], [202. , nan, 207.8, ..., nan, nan, nan], [ nan, nan, 198.1, ..., 11.1, 17.4, 28.3], ..., [348.6, 347.2, 316.2, ..., 209.5, 210.9, 212.7], [348.9, 320.1, 299.2, ..., 211.2, 212.2, 212.7], [334.2, 336.7, 368.1, ..., 210.6, 212. , 212.8]]) - oxygen_qc(N_PROF, N_LEVELS)float32nan 6.0 2.0 2.0 ... 2.0 2.0 2.0 2.0
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, 6., 2., ..., 2., 2., 2.], [ 2., nan, 2., ..., nan, nan, nan], [nan, nan, 2., ..., 2., 2., 2.], ..., [ 2., 2., 2., ..., 2., 2., 2.], [ 2., 2., 2., ..., 2., 2., 2.], [ 2., 2., 2., ..., 2., 2., 2.]], dtype=float32) - silicate(N_PROF, N_LEVELS)float64nan 1.5 2.3 ... 134.7 138.8 144.7
- whp_name :
- SILCAT
- whp_unit :
- UMOL/KG
- standard_name :
- moles_of_silicate_per_unit_mass_in_sea_water
- units :
- umol/kg
- C_format :
- %.1f
- C_format_source :
- input_file
- ancillary_variables :
- silicate_qc
array([[ nan, 1.5, 2.3, ..., 154.3, 158.1, 159.2], [ 1.9, nan, 2.1, ..., nan, nan, nan], [ nan, nan, 1.6, ..., 105.8, 115.4, 126.4], ..., [ 45.8, 49.2, 60.7, ..., 137. , 138.8, 143.8], [ 44.8, 60.9, 67.6, ..., 135.9, 139.6, 144.2], [ 25.6, 19.1, 21.6, ..., 134.7, 138.8, 144.7]]) - silicate_qc(N_PROF, N_LEVELS)float32nan 2.0 2.0 2.0 ... 6.0 6.0 2.0 2.0
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, 2., 2., ..., 6., 6., 6.], [ 2., nan, 2., ..., nan, nan, nan], [nan, nan, 2., ..., 2., 6., 6.], ..., [ 2., 2., 2., ..., 6., 2., 2.], [ 2., 2., 2., ..., 6., 2., 2.], [ 2., 2., 2., ..., 6., 2., 2.]], dtype=float32) - nitrate(N_PROF, N_LEVELS)float64nan 0.1 0.1 10.2 ... 33.1 33.2 33.3
- whp_name :
- NITRAT
- whp_unit :
- UMOL/KG
- standard_name :
- moles_of_nitrate_per_unit_mass_in_sea_water
- units :
- umol/kg
- C_format :
- %.1f
- C_format_source :
- input_file
- ancillary_variables :
- nitrate_qc
array([[ nan, 0.1, 0.1, ..., 39.9, 39.3, 38.9], [ 0. , nan, 0. , ..., nan, nan, nan], [ nan, nan, 0. , ..., 44.3, 44. , 44.6], ..., [24.8, 27.1, 29.6, ..., 33.2, 33.2, 33.3], [24.4, 28.8, 30.5, ..., 33.1, 33.2, 33.3], [22.9, 22.6, 23.6, ..., 33.1, 33.2, 33.3]]) - nitrate_qc(N_PROF, N_LEVELS)float32nan 2.0 2.0 2.0 ... 6.0 6.0 2.0 2.0
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, 2., 2., ..., 6., 6., 6.], [ 2., nan, 2., ..., nan, nan, nan], [nan, nan, 2., ..., 2., 6., 6.], ..., [ 2., 2., 2., ..., 6., 2., 2.], [ 2., 2., 2., ..., 6., 2., 2.], [ 2., 2., 2., ..., 6., 2., 2.]], dtype=float32) - nitrite(N_PROF, N_LEVELS)float64nan 0.01 0.03 ... 0.01 0.01 0.01
- whp_name :
- NITRIT
- whp_unit :
- UMOL/KG
- standard_name :
- moles_of_nitrite_per_unit_mass_in_sea_water
- units :
- umol/kg
- C_format :
- %.2f
- C_format_source :
- input_file
- ancillary_variables :
- nitrite_qc
array([[ nan, 0.01, 0.03, ..., 0.01, 0.01, 0.01], [0.02, nan, 0.01, ..., nan, nan, nan], [ nan, nan, 0.01, ..., 0.01, 0.01, 0.02], ..., [0.36, 0.18, 0.21, ..., 0.01, 0.01, 0.02], [0.36, 0.17, 0.19, ..., 0.02, 0.02, 0.02], [0.43, 0.36, 0.23, ..., 0.01, 0.01, 0.01]]) - nitrite_qc(N_PROF, N_LEVELS)float32nan 2.0 2.0 2.0 ... 6.0 6.0 2.0 2.0
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, 2., 2., ..., 6., 6., 6.], [ 2., nan, 2., ..., nan, nan, nan], [nan, nan, 2., ..., 2., 6., 6.], ..., [ 2., 2., 2., ..., 6., 2., 2.], [ 2., 2., 2., ..., 6., 2., 2.], [ 2., 2., 2., ..., 6., 2., 2.]], dtype=float32) - phosphate(N_PROF, N_LEVELS)float64nan 0.301 0.443 ... 2.275 2.272
- whp_name :
- PHSPHT
- whp_unit :
- UMOL/KG
- standard_name :
- moles_of_phosphate_per_unit_mass_in_sea_water
- units :
- umol/kg
- C_format :
- %.3f
- C_format_source :
- input_file
- ancillary_variables :
- phosphate_qc
array([[ nan, 0.301, 0.443, ..., 2.802, 2.758, 2.729], [0.287, nan, 0.269, ..., nan, nan, nan], [ nan, nan, 0.279, ..., 3.269, 3.265, 3.224], ..., [1.655, 1.921, 2.07 , ..., 2.271, 2.256, 2.27 ], [1.63 , 2.024, 2.123, ..., 2.264, 2.277, 2.275], [1.503, 1.521, 1.674, ..., 2.262, 2.275, 2.272]]) - phosphate_qc(N_PROF, N_LEVELS)float32nan 2.0 2.0 2.0 ... 6.0 6.0 2.0 2.0
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, 2., 2., ..., 6., 6., 6.], [ 2., nan, 2., ..., nan, nan, nan], [nan, nan, 2., ..., 2., 6., 6.], ..., [ 2., 2., 2., ..., 6., 2., 2.], [ 2., 2., 2., ..., 6., 2., 2.], [ 2., 2., 2., ..., 6., 2., 2.]], dtype=float32) - cfc_11(N_PROF, N_LEVELS)float64nan 1.514 2.099 ... nan nan nan
- whp_name :
- CFC-11
- whp_unit :
- PMOL/KG
- standard_name :
- moles_of_cfc11_per_unit_mass_in_sea_water
- units :
- pmol/kg
- C_format :
- %.4f
- C_format_source :
- input_file
- ancillary_variables :
- cfc_11_qc
array([[ nan, 1.5139, 2.0995, ..., nan, 0.009 , 0.0094], [1.6437, nan, 1.7631, ..., nan, nan, nan], [ nan, nan, 1.5528, ..., 0.0143, nan, 0.0169], ..., [6.0107, 5.4535, 5.0256, ..., 0.1906, 0.2211, 0.3853], [5.9748, 4.991 , 4.6783, ..., 0.2388, 0.2922, 0.4053], [ nan, nan, nan, ..., nan, nan, nan]]) - cfc_11_qc(N_PROF, N_LEVELS)float32nan 6.0 2.0 2.0 ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, 6., 2., ..., nan, 2., 2.], [ 6., nan, 2., ..., nan, nan, nan], [nan, nan, 2., ..., 2., nan, 2.], ..., [ 2., 2., 2., ..., 2., 2., 2.], [ 2., 2., 2., ..., 2., 2., 2.], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - cfc_12(N_PROF, N_LEVELS)float64nan 0.9507 1.267 ... nan nan nan
- whp_name :
- CFC-12
- whp_unit :
- PMOL/KG
- units :
- pmol/kg
- C_format :
- %.4f
- C_format_source :
- input_file
- ancillary_variables :
- cfc_12_qc
array([[ nan, 0.9507, 1.2668, ..., nan, 0.0063, 0.0063], [1.0138, nan, 1.08 , ..., nan, nan, nan], [ nan, nan, 0.9728, ..., 0.007 , nan, 0.0056], ..., [3.1982, 2.9252, 2.6889, ..., 0.0935, 0.1067, 0.1822], [3.2103, 2.6822, 2.5081, ..., 0.1145, 0.1406, 0.1929], [ nan, nan, nan, ..., nan, nan, nan]]) - cfc_12_qc(N_PROF, N_LEVELS)float32nan 6.0 2.0 2.0 ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, 6., 2., ..., nan, 2., 2.], [ 6., nan, 2., ..., nan, nan, nan], [nan, nan, 2., ..., 2., nan, 2.], ..., [ 2., 2., 2., ..., 2., 2., 2.], [ 2., 2., 2., ..., 2., 2., 2.], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - sulfur_hexifluoride(N_PROF, N_LEVELS)float64nan 1.36 1.666 ... nan nan nan
- whp_name :
- SF6
- whp_unit :
- FMOL/KG
- units :
- fmol/kg
- C_format :
- %.4f
- C_format_source :
- input_file
- ancillary_variables :
- sulfur_hexifluoride_qc
array([[ nan, 1.3599e+00, 1.6662e+00, ..., nan, 3.1000e-03, 0.0000e+00], [1.4978e+00, nan, 1.5242e+00, ..., nan, nan, nan], [ nan, nan, 1.4072e+00, ..., 3.2300e-02, nan, 0.0000e+00], ..., [3.6577e+00, 3.2482e+00, 2.9527e+00, ..., 3.7900e-02, 6.6300e-02, 7.1400e-02], [3.6171e+00, 2.9316e+00, 2.6959e+00, ..., 3.6000e-02, 4.4400e-02, 7.0100e-02], [ nan, nan, nan, ..., nan, nan, nan]]) - sulfur_hexifluoride_qc(N_PROF, N_LEVELS)float32nan 6.0 2.0 2.0 ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, 6., 2., ..., nan, 2., 2.], [ 6., nan, 2., ..., nan, nan, nan], [nan, nan, 2., ..., 2., nan, 2.], ..., [ 2., 2., 2., ..., 2., 2., 2.], [ 2., 2., 2., ..., 2., 2., 2.], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - total_carbon(N_PROF, N_LEVELS)float64nan 1.986e+03 ... 2.265e+03
- whp_name :
- TCARBN
- whp_unit :
- UMOL/KG
- standard_name :
- moles_of_dissolved_inorganic_carbon_per_unit_mass_in_sea_water
- units :
- umol/kg
- C_format :
- %.1f
- C_format_source :
- input_file
- ancillary_variables :
- total_carbon_qc
array([[ nan, 1985.9, 2040.5, ..., 2368.3, 2367.5, 2366.1], [1999.8, nan, 2008.6, ..., nan, nan, nan], [ nan, nan, 1986.4, ..., 2360.2, 2369.1, 2373.6], ..., [2083.5, 2161.2, 2194.4, ..., 2262.2, 2262.6, 2263.2], [2075.9, 2193.1, 2212.5, ..., 2263.9, 2263.4, 2264.1], [2102.5, 2126.5, 2132.9, ..., 2260.8, 2262.6, 2264.6]]) - total_carbon_qc(N_PROF, N_LEVELS)float32nan 6.0 2.0 2.0 ... 2.0 2.0 6.0 2.0
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, 6., 2., ..., 2., 2., 6.], [ 6., nan, 2., ..., nan, nan, nan], [nan, nan, 2., ..., 2., 2., 6.], ..., [ 6., 2., 2., ..., 2., 2., 6.], [ 6., 2., 2., ..., 2., 2., 6.], [ 6., 2., 2., ..., 2., 6., 2.]], dtype=float32) - total_alkalinity(N_PROF, N_LEVELS)float64nan 2.29e+03 ... 2.364e+03
- whp_name :
- ALKALI
- whp_unit :
- UMOL/KG
- units :
- umol/kg
- C_format :
- %.1f
- C_format_source :
- input_file
- ancillary_variables :
- total_alkalinity_qc
array([[ nan, 2289.5, 2278.6, ..., 2421.6, 2427.2, 2428.2], [2284.5, nan, 2270.2, ..., nan, nan, nan], [ nan, nan, 2289.4, ..., nan, 2377.4, 2389.2], ..., [2201.7, 2284.2, 2300.7, ..., 2366. , 2362. , 2362.7], [2189.8, 2298.8, 2309.4, ..., 2363.2, 2361.8, 2362.2], [2228.4, 2265.8, 2271.2, ..., 2361.6, 2362.2, 2363.6]]) - total_alkalinity_qc(N_PROF, N_LEVELS)float32nan 2.0 2.0 6.0 ... 6.0 2.0 2.0 2.0
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, 2., 2., ..., 6., 2., 2.], [ 6., nan, 2., ..., nan, nan, nan], [nan, nan, 6., ..., 5., 2., 2.], ..., [ 2., 2., 2., ..., 2., 2., 2.], [ 2., 2., 6., ..., 2., 6., 2.], [ 2., 2., 6., ..., 2., 2., 2.]], dtype=float32) - ph_sws(N_PROF, N_LEVELS)float64nan 8.056 7.95 ... 7.586 7.586
- whp_name :
- PH_SWS
- C_format :
- %.4f
- C_format_source :
- input_file
- ancillary_variables :
- ph_sws_qc ph_temperature
array([[ nan, 8.0557, 7.9498, ..., 7.4699, 7.4893, 7.5127], [8.028 , nan, 8.0039, ..., nan, nan, nan], [ nan, nan, 8.0479, ..., 7.3168, 7.3283, 7.3711], ..., [7.675 , 7.6684, 7.6152, ..., 7.5904, 7.588 , 7.5882], [7.676 , 7.6272, 7.5971, ..., 7.5895, 7.5867, 7.5843], [7.702 , 7.7181, 7.7148, ..., 7.588 , 7.5859, 7.5858]]) - ph_sws_qc(N_PROF, N_LEVELS)float32nan 2.0 6.0 2.0 ... 2.0 2.0 2.0 2.0
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, 2., 6., ..., 2., 2., 2.], [ 2., nan, 2., ..., nan, nan, nan], [nan, nan, 2., ..., 2., 2., 2.], ..., [ 2., 6., 2., ..., 2., 2., 2.], [ 2., 2., 2., ..., 2., 2., 6.], [ 2., 6., 2., ..., 2., 2., 2.]], dtype=float32) - ph_temperature(N_PROF, N_LEVELS)float64nan 25.0 25.0 ... 25.0 25.0 25.0
- whp_name :
- PH_TMP
- whp_unit :
- DEG C
- standard_name :
- temperature_of_analysis_of_sea_water
- units :
- degC
- reference_scale :
- unknown
- C_format :
- %.2f
- C_format_source :
- input_file
array([[nan, 25., 25., ..., 25., 25., 25.], [25., nan, 25., ..., nan, nan, nan], [nan, nan, 25., ..., 25., 25., 25.], ..., [25., 25., 25., ..., 25., 25., 25.], [25., 25., 25., ..., 25., 25., 25.], [25., 25., 25., ..., 25., 25., 25.]]) - dissolved_organic_carbon(N_PROF, N_LEVELS)float64nan nan nan nan ... nan nan nan nan
- whp_name :
- DOC
- whp_unit :
- UMOL/KG
- units :
- umol/kg
- C_format :
- %.1f
- C_format_source :
- input_file
- ancillary_variables :
- dissolved_organic_carbon_qc
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - dissolved_organic_carbon_qc(N_PROF, N_LEVELS)float32nan nan nan nan ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - del_carbon_13_dic(N_PROF, N_LEVELS)float64nan 1.27 1.07 0.5 ... nan nan nan
- whp_name :
- DELC13
- whp_unit :
- /MILLE
- units :
- 1e-3
- C_format :
- %.2f
- C_format_source :
- input_file
- ancillary_variables :
- del_carbon_13_dic_qc
array([[ nan, 1.27, 1.07, ..., -0.19, -0.33, -0.15], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - del_carbon_13_dic_qc(N_PROF, N_LEVELS)float32nan 2.0 2.0 2.0 ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, 2., 2., ..., 2., 2., 2.], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - del_carbon_14_dic(N_PROF, N_LEVELS)float64nan 11.4 7.5 5.6 ... nan nan nan
- whp_name :
- DELC14
- whp_unit :
- /MILLE
- units :
- 1e-3
- C_format :
- %.1f
- C_format_source :
- input_file
- ancillary_variables :
- del_carbon_14_dic_error del_carbon_14_dic_qc
array([[ nan, 11.4, 7.5, ..., -250.7, -245.6, -246.8], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - del_carbon_14_dic_qc(N_PROF, N_LEVELS)float32nan 2.0 2.0 2.0 ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, 2., 2., ..., 2., 2., 2.], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - del_carbon_14_dic_error(N_PROF, N_LEVELS)float64nan 2.6 2.8 2.7 ... nan nan nan nan
- whp_name :
- C14ERR
- whp_unit :
- /MILLE
- units :
- 1e-3
- C_format :
- %.1f
- C_format_source :
- input_file
array([[nan, 2.6, 2.8, ..., 2.1, 2.7, 2.3], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - particulate_organic_carbon(N_PROF, N_LEVELS)float64nan nan nan nan ... nan nan nan nan
- whp_name :
- POC
- whp_unit :
- UG/KG
- units :
- ug/kg
- C_format :
- %.2f
- C_format_source :
- input_file
- ancillary_variables :
- particulate_organic_carbon_qc
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - particulate_organic_carbon_qc(N_PROF, N_LEVELS)float32nan nan nan nan ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - particulate_organic_nitrogen(N_PROF, N_LEVELS)float64nan nan nan nan ... nan nan nan nan
- whp_name :
- PON
- whp_unit :
- UG/KG
- units :
- ug/kg
- C_format :
- %.2f
- C_format_source :
- input_file
- ancillary_variables :
- particulate_organic_nitrogen_qc
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - particulate_organic_nitrogen_qc(N_PROF, N_LEVELS)float32nan nan nan nan ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - total_dissolved_phosphorus(N_PROF, N_LEVELS)float64nan nan nan nan ... nan nan nan nan
- whp_name :
- TDP
- whp_unit :
- UMOL/KG
- units :
- umol/kg
- C_format :
- %.2f
- C_format_source :
- input_file
- ancillary_variables :
- total_dissolved_phosphorus_qc
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - total_dissolved_phosphorus_qc(N_PROF, N_LEVELS)float32nan nan nan nan ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - total_dissolved_nitrogen(N_PROF, N_LEVELS)float64nan nan nan nan ... nan nan nan nan
- whp_name :
- TDN
- whp_unit :
- UMOL/KG
- units :
- umol/kg
- C_format :
- %.2f
- C_format_source :
- input_file
- ancillary_variables :
- total_dissolved_nitrogen_qc
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - total_dissolved_nitrogen_qc(N_PROF, N_LEVELS)float32nan nan nan nan ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - carbon_tetrachloride(N_PROF, N_LEVELS)float64nan 1.274 0.9866 ... nan nan nan
- whp_name :
- CCL4
- whp_unit :
- PMOL/KG
- units :
- pmol/kg
- C_format :
- %.4f
- C_format_source :
- input_file
- ancillary_variables :
- carbon_tetrachloride_qc
array([[ nan, 1.2743, 0.9866, ..., nan, 0.12 , 0.0454], [1.5972, nan, 1.6429, ..., nan, nan, nan], [ nan, nan, 1.3544, ..., 0.0269, nan, 0.0448], ..., [0.3113, 0.3223, 0.3511, ..., 0.2615, 0.2503, 0.2937], [ nan, nan, nan, ..., 0.2651, 0.2287, 0.2885], [ nan, nan, nan, ..., nan, nan, nan]]) - carbon_tetrachloride_qc(N_PROF, N_LEVELS)float32nan 2.0 2.0 2.0 ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, 2., 2., ..., nan, 2., 2.], [ 2., nan, 2., ..., nan, nan, nan], [nan, nan, 2., ..., 2., nan, 2.], ..., [ 2., 2., 2., ..., 2., 2., 2.], [ 5., 5., 5., ..., 2., 2., 2.], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - nitrous_oxide(N_PROF, N_LEVELS)float64nan 6.393 15.24 ... nan nan nan
- whp_name :
- N2O
- whp_unit :
- NMOL/KG
- units :
- nmol/kg
- C_format :
- %.4f
- C_format_source :
- input_file
- ancillary_variables :
- nitrous_oxide_qc
array([[ nan, 6.3926, 15.2363, ..., nan, 24.9708, 24.4458], [ 6.4918, nan, 6.8435, ..., nan, nan, nan], [ nan, nan, 6.6349, ..., 52.1009, nan, 43.9071], ..., [15.6019, 17.0105, 17.8368, ..., 18.2111, 18.5337, 18.4587], [15.4805, 17.5179, 17.8503, ..., 18.2908, 18.0859, 18.2292], [ nan, nan, nan, ..., nan, nan, nan]]) - nitrous_oxide_qc(N_PROF, N_LEVELS)float32nan 2.0 2.0 2.0 ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, 2., 2., ..., nan, 2., 2.], [ 2., nan, 2., ..., nan, nan, nan], [nan, nan, 2., ..., 2., nan, 2.], ..., [ 2., 2., 2., ..., 2., 2., 2.], [ 2., 2., 2., ..., 2., 2., 2.], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - nitrous_oxide_l_alt_1(N_PROF, N_LEVELS)float64nan nan nan nan ... nan nan nan nan
- whp_name :
- N2O_ALT_1
- whp_unit :
- NMOL/L
- units :
- nmol/l
- C_format :
- %.9f
- C_format_source :
- input_file
- ancillary_variables :
- nitrous_oxide_l_alt_1_qc
array([[ nan, nan, nan, ..., nan, nan, nan], [6.89973917, nan, 7.1366651 , ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - nitrous_oxide_l_alt_1_qc(N_PROF, N_LEVELS)float32nan nan nan nan ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, nan, nan, ..., nan, nan, nan], [ 2., nan, 2., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - dissolved_organic_carbon_14(N_PROF, N_LEVELS)float64nan nan nan nan ... nan nan nan nan
- whp_name :
- 14C-DOC
- whp_unit :
- /MILLE
- units :
- 1e-3
- C_format :
- %.1f
- C_format_source :
- input_file
- ancillary_variables :
- dissolved_organic_carbon_14_error dissolved_organic_carbon_14_qc
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - dissolved_organic_carbon_14_qc(N_PROF, N_LEVELS)float32nan nan nan nan ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - dissolved_organic_carbon_14_error(N_PROF, N_LEVELS)float64nan nan nan nan ... nan nan nan nan
- whp_name :
- 14C-DOCERR
- whp_unit :
- /MILLE
- units :
- 1e-3
- C_format :
- %.1f
- C_format_source :
- input_file
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - dissolved_organic_carbon_13(N_PROF, N_LEVELS)float64nan nan nan nan ... nan nan nan nan
- whp_name :
- 13C-DOC
- whp_unit :
- /MILLE
- units :
- 1e-3
- C_format :
- %.1f
- C_format_source :
- input_file
- ancillary_variables :
- dissolved_organic_carbon_13_qc
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - dissolved_organic_carbon_13_qc(N_PROF, N_LEVELS)float32nan nan nan nan ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - d15n_no3(N_PROF, N_LEVELS)float64nan nan nan nan ... nan nan nan nan
- whp_name :
- D15N_NO3
- whp_unit :
- /MILLE
- units :
- 1e-3
- C_format :
- %.2f
- C_format_source :
- input_file
- ancillary_variables :
- d15n_no3_error d15n_no3_qc
array([[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [7.31, 6.02, 5.47, ..., 4.68, 4.62, 4.74], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - d15n_no3_qc(N_PROF, N_LEVELS)float32nan nan nan nan ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [ 2., 2., 2., ..., 2., 2., 2.], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - d15n_no3_error(N_PROF, N_LEVELS)float64nan nan nan nan ... nan nan nan nan
- whp_name :
- D15N_NO3_ERROR
- whp_unit :
- /MILLE
- units :
- 1e-3
- C_format :
- %.2f
- C_format_source :
- input_file
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - d15n_n2o(N_PROF, N_LEVELS)float64nan nan nan nan ... nan nan nan nan
- whp_name :
- D15N_N2O
- whp_unit :
- /MILLE
- units :
- 1e-3
- C_format :
- %.2f
- C_format_source :
- input_file
- ancillary_variables :
- d15n_n2o_qc
array([[ nan, nan, nan, ..., nan, nan, nan], [8.34, nan, 8.08, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - d15n_n2o_qc(N_PROF, N_LEVELS)float32nan nan nan nan ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, nan, nan, ..., nan, nan, nan], [ 2., nan, 2., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - d15n_alpha_n2o(N_PROF, N_LEVELS)float64nan nan nan nan ... nan nan nan nan
- whp_name :
- D15N_ALPHA_N2O
- whp_unit :
- /MILLE
- units :
- 1e-3
- C_format :
- %.2f
- C_format_source :
- input_file
- ancillary_variables :
- d15n_alpha_n2o_qc
array([[ nan, nan, nan, ..., nan, nan, nan], [21.53, nan, 21.08, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - d15n_alpha_n2o_qc(N_PROF, N_LEVELS)float32nan nan nan nan ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, nan, nan, ..., nan, nan, nan], [ 2., nan, 2., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - d15n_nitrite_nitrate(N_PROF, N_LEVELS)float64nan nan nan nan ... nan nan nan nan
- whp_name :
- D15N_NO2+NO3
- whp_unit :
- /MILLE
- units :
- 1e-3
- C_format :
- %.2f
- C_format_source :
- input_file
- ancillary_variables :
- d15n_nitrite_nitrate_qc
array([[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [6.08, 5.71, 5.18, ..., 4.68, 4.62, 4.74], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - d15n_nitrite_nitrate_qc(N_PROF, N_LEVELS)float32nan nan nan nan ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [ 2., 2., 2., ..., 2., 2., 2.], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - d18o_nitrite_nitrate(N_PROF, N_LEVELS)float64nan nan nan nan ... nan nan nan nan
- whp_name :
- D18O_NO2+NO3
- whp_unit :
- /MILLE
- units :
- 1e-3
- C_format :
- %.2f
- C_format_source :
- input_file
- ancillary_variables :
- d18o_nitrite_nitrate_qc
array([[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [2.96, 2.66, 2.47, ..., 1.67, 1.61, 1.82], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - d18o_nitrite_nitrate_qc(N_PROF, N_LEVELS)float32nan nan nan nan ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [ 2., 2., 2., ..., 2., 2., 2.], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - d18o_nitrate(N_PROF, N_LEVELS)float64nan nan nan nan ... nan nan nan nan
- whp_name :
- D18O_NO3
- whp_unit :
- /MILLE
- units :
- 1e-3
- C_format :
- %.2f
- C_format_source :
- input_file
- ancillary_variables :
- d18o_nitrate_qc
array([[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [2.85, 2.75, 2.4 , ..., 1.67, 1.61, 1.82], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - d18o_nitrate_qc(N_PROF, N_LEVELS)float32nan nan nan nan ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [ 2., 2., 2., ..., 2., 2., 2.], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - d18o_nitrust_oxide(N_PROF, N_LEVELS)float64nan nan nan nan ... nan nan nan nan
- whp_name :
- D18O_N2O
- whp_unit :
- /MILLE
- units :
- 1e-3
- C_format :
- %.2f
- C_format_source :
- input_file
- ancillary_variables :
- d18o_nitrust_oxide_qc
array([[ nan, nan, nan, ..., nan, nan, nan], [54.18, nan, 52.61, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - d18o_nitrust_oxide_qc(N_PROF, N_LEVELS)float32nan nan nan nan ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, nan, nan, ..., nan, nan, nan], [ 2., nan, 2., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - n2_argon_ratio(N_PROF, N_LEVELS)float64nan nan nan nan ... nan nan nan nan
- whp_name :
- N2/ARGON
- C_format :
- %.2f
- C_format_source :
- input_file
- ancillary_variables :
- n2_argon_ratio_qc n2_argon_ratio_error
array([[ nan, nan, nan, ..., nan, nan, nan], [38.36, nan, 38.18, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - n2_argon_ratio_qc(N_PROF, N_LEVELS)float32nan nan nan nan ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, nan, nan, ..., nan, nan, nan], [ 2., nan, 2., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - n2_argon_ratio_error(N_PROF, N_LEVELS)float64nan nan nan nan ... nan nan nan nan
- whp_name :
- N2/ARGON_ERROR
- C_format :
- %.2f
- C_format_source :
- input_file
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - n2_argon_ratio_unstripped(N_PROF, N_LEVELS)float64nan nan nan nan ... nan nan nan nan
- whp_name :
- N2/ARGON_UNSTRIPPED
- C_format :
- %.2f
- C_format_source :
- input_file
- ancillary_variables :
- n2_argon_ratio_unstripped_error n2_argon_ratio_unstripped_qc
array([[ nan, nan, nan, ..., nan, nan, nan], [38.37, nan, 38.25, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - n2_argon_ratio_unstripped_qc(N_PROF, N_LEVELS)float32nan nan nan nan ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, nan, nan, ..., nan, nan, nan], [ 2., nan, 2., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - n2_argon_ratio_unstripped_error(N_PROF, N_LEVELS)float64nan nan nan nan ... nan nan nan nan
- whp_name :
- N2/ARGON_UNSTRIPPED_ERROR
- C_format :
- %.2f
- C_format_source :
- input_file
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - d15n_n2(N_PROF, N_LEVELS)float64nan nan nan nan ... nan nan nan nan
- whp_name :
- D15N_N2
- whp_unit :
- /MILLE
- units :
- 1e-3
- C_format :
- %.2f
- C_format_source :
- input_file
- ancillary_variables :
- d15n_n2_error d15n_n2_qc
array([[ nan, nan, nan, ..., nan, nan, nan], [0.59, nan, 0.43, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - d15n_n2_qc(N_PROF, N_LEVELS)float32nan nan nan nan ... nan nan nan nan
- standard_name :
- status_flag
- flag_values :
- [0 1 2 3 4 5 6 7 8 9]
- flag_meanings :
- no_flag_assigned sample_for_this_measurement_was_drawn_from_water_bottle_but_analysis_not_received acceptable_measurement questionable_measurement bad_measurement not_reported mean_of_replicate_measurements manual_chromatographic_peak_measurement irregular_digital_chromatographic_peak_integration sample_not_drawn_for_this_measurement_from_this_bottle
- conventions :
- WOCEBOTTLE - WOCE Quality Codes for water sample (bottle) measurements
array([[nan, nan, nan, ..., nan, nan, nan], [ 3., nan, 2., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - d15n_n2_error(N_PROF, N_LEVELS)float64nan nan nan nan ... nan nan nan nan
- whp_name :
- D15N_N2_ERROR
- whp_unit :
- /MILLE
- units :
- 1e-3
- C_format :
- %.2f
- C_format_source :
- input_file
array([[nan, nan, nan, ..., nan, nan, nan], [0.1, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - profile_type(N_PROF)object'B' 'B' 'B' 'B' ... 'B' 'B' 'B' 'B'
array(['B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B', 'B'], dtype=object) - geometry_container()float64nan
- geometry_type :
- point
- node_coordinates :
- longitude latitude
array(nan)
- Conventions :
- CF-1.8 CCHDO-1.0
- cchdo_software_version :
- hydro 1.0.2.8
- cchdo_parameters_version :
- params 2024.4.0
- comments :
- BOTTLE,20230606CCHSIOCBG Merged parameters: N2O_ALT_1, N2O_ALT_1_FLAG_W, D15N_N2O, D15N_N2O_FLAG_W, D18O_N2O, D18O_N2O_FLAG_W, D15N_ALPHA_N2O, D15N_ALPHA_N2O_FLAG_W Merged parameters: TDP, TDP_FLAG_W 20211118CCHSIOCBG Merged parameters: D15N_NO2+NO3, D15N_NO2+NO3_FLAG_W, D18O_NO2+NO3, D18O_NO2+NO3_FLAG_W, D15N_NO3, D15N_NO3_FLAG_W, D18O_NO3, D18O_NO3_FLAG_W Merged parameters: N2/ARGON_UNSTRIPPED, N2/ARGON_UNSTRIPPED_ERROR, N2/ARGON_UNSTRIPPED_FLAG_W, N2/ARGON, N2/ARGON_ERROR, N2/ARGON_FLAG_W, D15N_N2, D15N_N2_ERROR, D15N_N2_FLAG_W, D15N_NO3, D15N_NO3_ERROR, D15N_NO3_FLAG_W Merged parameters: N2O, N2O_FLAG_W, CCL4, CCL4_FLAG_W Merged parameters: 14C-DOC, 14C-DOC_FLAG_W, 14C-DOCERR, 13C-DOC, 13C-DOC_FLAG_W Merged parameters: DELC13, DELC13_FLAG_W, DELC14, DELC14_FLAG_W, C14ERR Merged parameters: DOC, DOC_FLAG_W, TDN, TDN_FLAG_W 20180921CCHSIOCBG Merged parameters: CTDPRS, CTDTMP, CTDSAL, SALNTY, SALNTY_FLAG_W, CTDOXY, CTDOXY_FLAG_W 20180103CCHSIOCBG Merged parameters: PHSPHT, PHSPHT_FLAG_W, SILCAT, SILCAT_FLAG_W, NITRAT, NITRAT_FLAG_W, NITRIT, NITRIT_FLAG_W Merged parameters: CFC-11, CFC-11_FLAG_W, CFC-12, CFC-12_FLAG_W, SF6, SF6_FLAG_W Merged parameters: OXYGEN_FLAG_W Merged parameters: ALKALI, ALKALI_FLAG_W, PH_SWS, PH_SWS_FLAG_W, PH_TMP Merged parameters: TCARBN, TCARBN_FLAG_W 20170505PRINUNIVRMK processed by CCHDO on 2017-05-15, units for POC/PON edited to UG/KG 20170807PRINUNIVRMK BOTTLE,20210212PRINUNIVRMK 06/06/2016 Initialized README file Data source: S.Purkey 2/8/17 SHIP: Ron Brown Cruise: P18; RB-16-06 Region: SE Pacific on 110W and 103W EXPOCODE: 33RO20161119 DATES:Leg 1: 20161119 - 20161223; San Diego to Easter Island Leg 2: 20161230 - 20170203; Easter Island to Punta Arenas Chief Scientist: Brendan Carter; Annie Bourbonnais - Leg 1 Rolf Sonnerup; Sarah Purkey - Leg 2 212 stations with 24 place Rosette SOCCOM Biogeochemical floats deployed by Sta WMO_ID Lat Lon Date UW# 128 5904844 -29.9815 -103.0012 20170104 9766 138 5904843 -34.9995 -102.9975 20170106 9659 146 5904685 -38.9988 -103.0015 20170108 9642 158 5904841 -44.9912 -102.9998 20170112 12382 170 5904842 -50.9990 -103.0005 20170115 12552 186 5904845 -58.9990 -102.9988 20170120 9750 202 5905075 -66.9960 -102.9992 20170124 8501 Supported by NSF Award PLR-1425989 to J.L. Sarmiento et al. Extra stations executed near SOCCOM floats from earlier cruises for cliberation Station/Cast Date Lat Lon Expocode/Sect/Sta/Cast UW# WMO_ID 195/1 20170122 -63.5163 -102.9805 320620140320/P16S/17/2 9092 5904185 212/1 20170129 -68.0655 -95.0013 320620161224/HazMat/16/2 12559 5904855 CTD: Who - G.Johnson; M.Barringer; Status - Includes: CTDSAL, CTDOXY, Optical XMISS: Who - W.Gardner; Status - final Notes: FLBB: Who - E.Boss; Status - final Notes: Salinty: Who - M.Barringer; Status - final Notes: Nutrients: Who - C.Mordy;J.-Z. Zhang; Status - final Includes: nitrate, nitrite, silicate, phopshpate O2: Who - C.Langdon; Status - final Notes: revised data and flags rcd 3/16/17. Replaced 3/17/17 Some additional flag changes possible TCO2: Who - R.Feely & R.Wannikohf; Status - final Notes: preliminary data from C.Featherstone via J.Bullister 3/7/17 final data from D.Greeley 8/2/17 Alkalinity: Who - F.Millero; Status - final Notes: data from R.Woosley via Kozyr 8/3/17 Additional QC by Key pH: Who - F.Millero; Status - final Notes: data from R.Woosley via Kozyr 8/3/17 Additional QC by Key NOTE: Data are on SWS pCO2: Who - R.Wannikohf; Status - Notes: underway only CFC: Who - J.Bullister; Status - final Notes: includes SF6 CCL4 measured, but not ready to report Final data submitted 9/19/17 C14/C13: Who - R.Key,A.McNichol; Status - final Notes: Final data with QC added 7/27/20 DO14C/DO13C: Who - E.Druffel; Status - final Notes: Final data with QC added 2/12/21 H3/He3: Who - S.Doney; Status - collected Notes: HPLC: Who - E.Boss; Status - submitted to NASA for analysis Notes: POC/PON: Who - E.Boss; Status - final Notes: collected by C.Kovach; final data merged 20170505 and units converted to ug/kg Approximate detection limits: POC - 0.5microgram/L PON - 3.4microgram/L TDN/DOC: Who - D.Hansell; Status - final Notes: merged with minor QC by Key, 12/20/18 Nitrogen isomers: Who - D.Sigman; Status - collected Notes: REE: Who - ?; Status - collected Notes: These data should be cited as: References that have used data from this cruise: DATA CITATION | CCHDO Citation Information | | Please cite data downloaded from CCHDO as: | [Data providers of the parameter(s) used]. [Year of file access]. [CTD/Bottle] data from cruise [expocode], | [format version used]. Accessed from CCHDO [url of cruise data page]. Access date [date of download]. | [Applicable CCHDO cruise DOI if provided]. | | For example: | Swift, J. and Becker, S. 2019. CTD data from Cruise 33RR20160321, exchange version. Accessed from CCHDO | https://cchdo.ucsd.edu/cruise/33RR20160321. Access date 2019-08-21. CCHDO cruise DOI: 10.7942/C2008W | | Information on the scientists who provided the different parameters can be found in the cruise report. | Additionally, for US GO-SHIP cruises, this information is usually in the comments section in the text headers | of the Exchange bottle files. | | Users are also encouraged to acknowledge the funding program that supported the data collection in their publication. | For US GO-SHIP data this is the NSF/NOAA funded U.S Repeat Hydrography Program. | | For data citation, please use the data providers listed below. | | Programs and Principal Investigators | | Program PI Inst. E-mail | ------------------- ----------------- ------- -------------------------- | CTD Greg Johnson PMEL gregory.c.johnson@noaa.gov | Molly Baringer AOML Molly.Baringer@noaa.gov | Salinity Molly Baringer AOML Molly.Baringer@noaa.gov | LADCP Andreas Thurnherr LDEO ant@ldeo.columbia.edu | Dissolved O2 Chris Langdon RSMAS clangdon@rsmas.miami.edu | Nutrients Calvin Mordy PMEL Calvin.W.Mordy@noaa.gov | Jia-Zhong Zhang AOML Jia-Zhong.Zhang@noaa.gov | CFCs/SF6/N2O by ECD John Bullister PMEL John.L.Bullister@noaa.gov | Total CO2 (DIC) Richard Feely PMEL richard.a.feely@noaa.gov | Rik Wannikhof AOML Rik.Wanninkhof@noaa.gov | Total Alkalinity/pH Frank Millero RSMAS fmillero@rsmas.miami.edu | DI14C Ann McNichol WHOI amcnichol@whoi.edu | Robert Key PU 401 Sayre Hall | Princeton University | 08544 Princeton , NJ | DO14C Ellen Druffel UCI edruffel@uci.edu | Dissolved Organics Brett Walker UCI Brett.walker@uci.edu | DOC and TDN Dennis Hansell RSMAS dhansell@rsmas.miami.edu | Helium/Tritium Scott Doney WHOI sdoney@whoi.edu | (3He-3H) William Jenkins WHOI wjenkins@whoi.edu | Stable Gases, O2, Annie Bourbonnais WHOI abourbonnais@whoi.edu | N2, Ar | Floats Greg Johnson PMEL Gregory.C.Johnson@noaa.gov | Lynne Talley SIO ltalley@ucsd.edu | Craig McNeil UW cmcneil@apl.washington.edu | Steve Emerson UW emerson@u.washington.edu | Giorgio Dall'Olmo PML gdal@pml.ac.uk | Drifters Shaun Dolk AOML shaun.dolk@noaa.gov | HPLC Emmanuel Boss UMe emmanuel.boss@maine.edu | Transmissometry Wilf Gardner TAMU wgardner@ocean.tamu.edu | Fluor./backscatter Emmanuel Boss UMe emmanuel.boss@maine.edu | Chipods Jonathan Nash OSU nash.coas@gmail.com | Net tows* Laura Sanchez CICIMAR lsvelasc@gmail.com | Velasco | Bathymetry Ship personnel NOAA ops.ronald.brown@noaa.gov | Underway TSG Ship personnel NOAA ops.ronald.brown@noaa.gov | Underway pCO2 Rik Wanninkhof AOML Rik.Wanninkhof@noaa.gov | Genetics Adam Martiny UCI amartiny@uci.edu | iTag Genetics Eric Allen SIO eallen@ucsd.edu | Bethany Kolody SIO bck243@gmail.com | Nitrate/nitrite Daniel Sigman PU sigman@princeton.edu | isotopes | FranÃÂçois Fripiat MPIC f.fripiat@mpic.de | Annie Bourbonnais WHOI abourbonnais@whoi.edu | N2O isotopomers Annie Bourbonnais WHOI abourbonnais@whoi.edu | Radiometry/ Michael Ondrusek NESDIS Michael.Ondrusek@noaa.gov | Chlorophyll | H2O isotopes Kim Cobb GIT kim.cobb@eas.gatech.edu | DO14C by Pyrolysis Lihini Aluwihare SIO laluwihare@ucsd.edu | Microplastics Shaowu Bao CCCU sbao@coastal.edu | ___________________________________________________________________________ | * Program cancelled after the first cast
- featureType :
- profile
# List the variables in this bottle file
all_vars = [i for i in dd.data_vars]
print(all_vars)
['section_id', 'bottle_number', 'bottle_number_qc', 'btm_depth', 'ctd_temperature', 'ctd_salinity', 'ctd_salinity_qc', 'bottle_salinity', 'bottle_salinity_qc', 'ctd_oxygen', 'ctd_oxygen_qc', 'oxygen', 'oxygen_qc', 'silicate', 'silicate_qc', 'nitrate', 'nitrate_qc', 'nitrite', 'nitrite_qc', 'phosphate', 'phosphate_qc', 'cfc_11', 'cfc_11_qc', 'cfc_12', 'cfc_12_qc', 'sulfur_hexifluoride', 'sulfur_hexifluoride_qc', 'total_carbon', 'total_carbon_qc', 'total_alkalinity', 'total_alkalinity_qc', 'ph_sws', 'ph_sws_qc', 'ph_temperature', 'dissolved_organic_carbon', 'dissolved_organic_carbon_qc', 'del_carbon_13_dic', 'del_carbon_13_dic_qc', 'del_carbon_14_dic', 'del_carbon_14_dic_qc', 'del_carbon_14_dic_error', 'particulate_organic_carbon', 'particulate_organic_carbon_qc', 'particulate_organic_nitrogen', 'particulate_organic_nitrogen_qc', 'total_dissolved_phosphorus', 'total_dissolved_phosphorus_qc', 'total_dissolved_nitrogen', 'total_dissolved_nitrogen_qc', 'carbon_tetrachloride', 'carbon_tetrachloride_qc', 'nitrous_oxide', 'nitrous_oxide_qc', 'nitrous_oxide_l_alt_1', 'nitrous_oxide_l_alt_1_qc', 'dissolved_organic_carbon_14', 'dissolved_organic_carbon_14_qc', 'dissolved_organic_carbon_14_error', 'dissolved_organic_carbon_13', 'dissolved_organic_carbon_13_qc', 'd15n_no3', 'd15n_no3_qc', 'd15n_no3_error', 'd15n_n2o', 'd15n_n2o_qc', 'd15n_alpha_n2o', 'd15n_alpha_n2o_qc', 'd15n_nitrite_nitrate', 'd15n_nitrite_nitrate_qc', 'd18o_nitrite_nitrate', 'd18o_nitrite_nitrate_qc', 'd18o_nitrate', 'd18o_nitrate_qc', 'd18o_nitrust_oxide', 'd18o_nitrust_oxide_qc', 'n2_argon_ratio', 'n2_argon_ratio_qc', 'n2_argon_ratio_error', 'n2_argon_ratio_unstripped', 'n2_argon_ratio_unstripped_qc', 'n2_argon_ratio_unstripped_error', 'd15n_n2', 'd15n_n2_qc', 'd15n_n2_error', 'profile_type', 'geometry_container']
exclude_vars = ['bottle_number'] # exclude these variables
# get a list of variables that we might be interested in-
# Filter remove one dimensional variables, QC flags and error variables
vars_of_interest = [i for i in all_vars if '_qc' not in i and '_error' not in i
and i not in exclude_vars
and len(dd[i].dims)==2]
print(vars_of_interest)
['ctd_temperature', 'ctd_salinity', 'bottle_salinity', 'ctd_oxygen', 'oxygen', 'silicate', 'nitrate', 'nitrite', 'phosphate', 'cfc_11', 'cfc_12', 'sulfur_hexifluoride', 'total_carbon', 'total_alkalinity', 'ph_sws', 'ph_temperature', 'dissolved_organic_carbon', 'del_carbon_13_dic', 'del_carbon_14_dic', 'particulate_organic_carbon', 'particulate_organic_nitrogen', 'total_dissolved_phosphorus', 'total_dissolved_nitrogen', 'carbon_tetrachloride', 'nitrous_oxide', 'nitrous_oxide_l_alt_1', 'dissolved_organic_carbon_14', 'dissolved_organic_carbon_13', 'd15n_no3', 'd15n_n2o', 'd15n_alpha_n2o', 'd15n_nitrite_nitrate', 'd18o_nitrite_nitrate', 'd18o_nitrate', 'd18o_nitrust_oxide', 'n2_argon_ratio', 'n2_argon_ratio_unstripped', 'd15n_n2']
# Calculate Depth as a function of pressure, latitude
depth = gsw.z_from_p(dd.pressure,
dd.latitude,)
We can plot some of these variables of interest in a subplot together#
Lets plot 16 variables in a 4x4 grid
Select a profile number profile_num to plot these 25 variables from
Select some plotting colors
Loop through these and plot
num_plots = 16 # number of variables to plot
profile_num = 190 # select profile number to plot
# Choose the colormap
cmap = mpl.colormaps['Spectral']
# Get evenly spaced colors from the colormap
colors = [cmap(i) for i in np.linspace(0, 256, num_plots, dtype=int)]
num_rows = int(np.sqrt(num_plots))
num_cols = int(np.sqrt(num_plots))
fig, axs = plt.subplots(num_rows, num_cols, figsize=(10, 10),sharey=True)
# Loop through variables and plot in each subplot
for i, var in enumerate(vars_of_interest[:16]):
# Calculate subplot index
row = i // num_rows
col = i % num_cols
# Plot variable on the corresponding subplot
axs[row, col].plot(dd[var][profile_num],depth[profile_num],
label=var,linewidth='4',color=colors[i])
axs[row, col].set_title(var)
axs[row, col].set_ylabel('Depth [m]')
plt.tight_layout() # Adjust layout
plt.show() # Show the plot
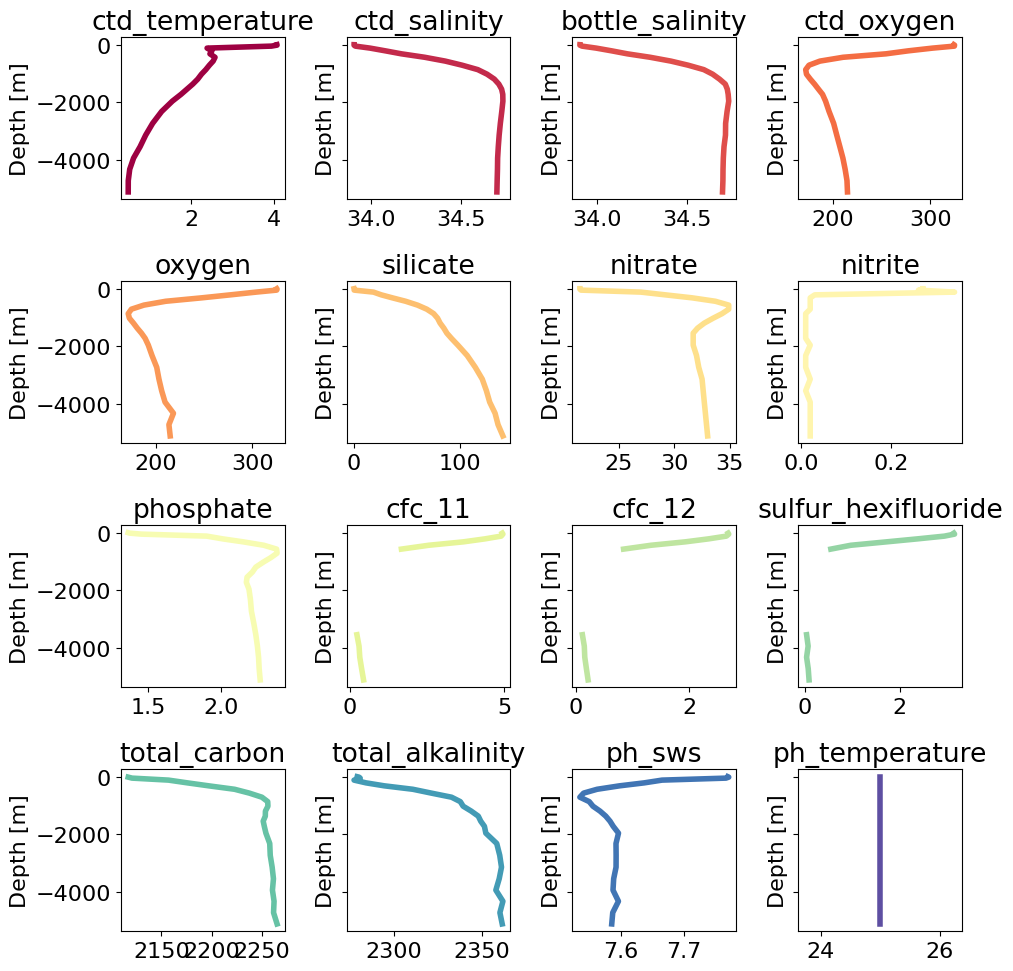
Next, we can calculate Absolute Salinity (SA) and Conservative Temperature (\(\Theta\)) and Potential Density (\(\sigma_0\))using the GSW Toolbox#
dd['SA'] = gsw.SA_from_SP(dd.ctd_salinity, dd.pressure, dd.longitude, dd.latitude)
dd['CT'] = gsw.CT_from_t(dd.SA,dd.ctd_temperature, dd.pressure)
dd['sigma_0'] = gsw.sigma0(dd.SA,dd.CT)
Now we can plot a T-S diagram using this information for all profiles along the section.#
We will also contour potential density, which is useful to identify water masses and water-mass transformation
plt.figure(figsize=(4,3),dpi=200)
# Create a grid of SA and CT values using meshgrid
SA_vals = np.linspace(dd.SA.min().item(), dd.SA.max().item(), 100)
CT_vals = np.linspace(dd.CT.min().item(), dd.CT.max().item(), 100)
SA_grid, CT_grid = np.meshgrid(SA_vals, CT_vals)
# Calculate potential density at each point on the grid
p_density = gsw.sigma0(SA_grid, CT_grid) # Assuming reference pressure of 0 dbar
# Scatter plot of Absolute Salinity vs. Conservative Temperature
plt.plot(dd.SA, dd.CT,
color='slateblue',
marker='+',
markersize=1.2,
alpha=0.8,
linestyle='')
# Add potential density contours
density_levels = np.linspace(min(p_density.flatten()), max(p_density.flatten()), 10) # Choose density levels
CS = plt.contour(SA_grid, CT_grid, p_density,
levels=density_levels,
colors='gray',
linestyles='dashed')
plt.clabel(CS, inline=True, fontsize=6, fmt='%.1f')
# Label axes
plt.xlabel('Absolute Salinity [g/kg]')
plt.ylabel(r'$\Theta$ [$^\circ$C]')
# Set axis limit
plt.xlim(32,36.6)
plt.show()